Cancer immunotherapy has transformed cancer treatment, but the complex tumor microenvironment (TME) hinders its effectiveness. Enter scRNA-seq, a revolutionary tool dissecting the TME at an unprecedented level.
Why scRNA-seq Matters
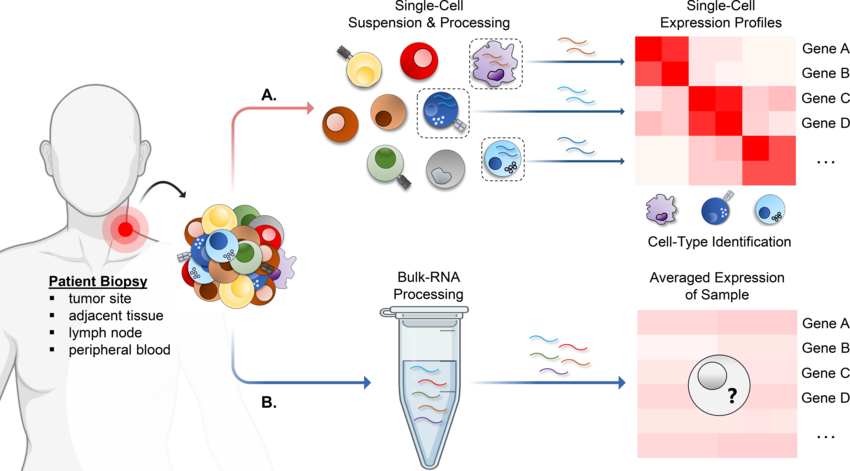
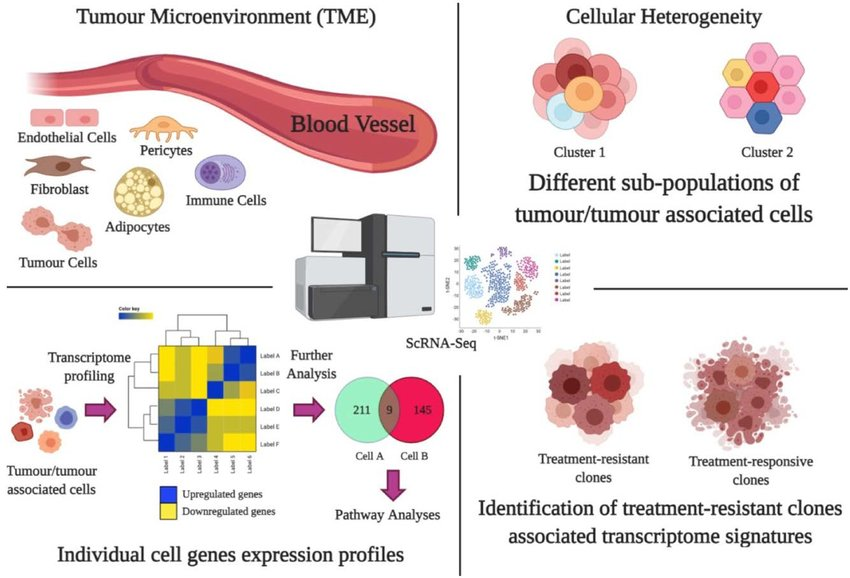
Traditional bulk RNA-seq averages gene expression across all cells, masking the TME's critical heterogeneity. The TME is a dynamic ecosystem teeming with diverse cell types, each playing a distinct role in the immune response to cancer.
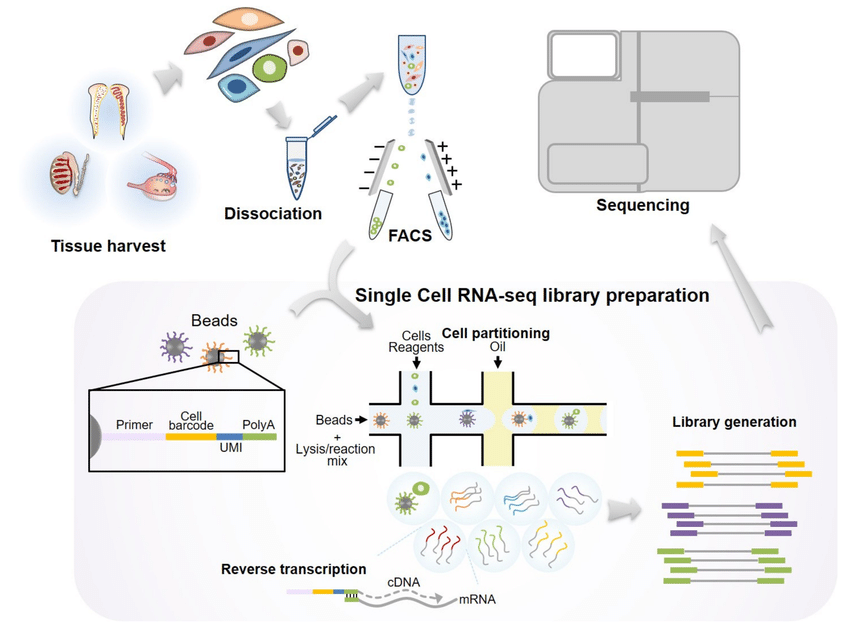
scRNA-seq breaks this barrier by analyzing gene expression at the single-cell level. This allows researchers to identify and characterize rare cell populations within the TME, such as functionally distinct subpopulations of immune cells. Imagine uncovering a hidden army within the tumor, some allies (immune cells) and some foes (suppressive cells). scRNA-seq equips us to understand their roles.
Considering a research project in this area? Maxanim can be your supplier for all component needs.
The Power of scRNA-seq in Immunotherapy Research
- Personalized Treatment with Biomarkers: scRNA-seq can identify gene signatures associated with response or resistance to specific immunotherapies. This paves the way for personalized treatment strategies, predicting which patients will benefit most from a particular immunotherapy.
- Discovering Novel Therapeutic Targets: By analyzing immunosuppressive cells, scRNA-seq helps identify novel therapeutic targets. Targeting these cells or their suppressive pathways could significantly enhance immunotherapy efficacy.
- Deciphering Cellular Communication: scRNA-seq allows for reconstructing intercellular communication networks within the TME. This reveals how different cell types interact and influence each other's function, providing insights into how tumors evade the immune system.
The Future of scRNA-seq in Immunotherapy
scRNA-seq is rapidly evolving, holding immense potential to advance cancer immunotherapy research. Challenges remain, such as analyzing the vast amount of data and integrating scRNA-seq with other modalities like spatial transcriptomics to understand the TME's spatial organization.
Conclusion
scRNA-seq is transforming our understanding of the TME, offering a window into the intricate cellular interactions that govern the immune response to cancer. By harnessing this power, researchers are poised to develop more targeted and effective immunotherapies, ultimately improving patient outcomes.
This video explains single-cell sequencing in 2 minutes
scRNA-seq: Unlocking the TME for Immunotherapy